Despite large increases in the uptake of screening in the past two decades, colorectal cancer (CRC) is still the second leading cause of cancer death in the US. This points to inadequate screening and treatment, and gaps in care that need to be addressed. Technological advances are bringing new methods for risk-targeted screening and treatment and new screening modalities. There is a critical need to assess these potential improvements in terms of both their ability to reduce the burden of CRC and their associated costs. But it is not logistically or ethically feasible to conduct clinical trials of all possible interventions. Instead, computational models, in the form of natural history microsimulations, are used as in silico laboratories to evaluate the potential impact of changes in clinical practice and new policies on clinical and economic CRC outcomes. These models are based on information about underlying disease process, sensitivity and specificity of screening tests, and treatment effectiveness. There is uncertainty in available data, which is observed with error, and the models, which describe unobservable processes. In the face of these uncertainties, large-scale computation is required to provide robust evidence for effective screening approaches.
This project will use leadership class computing resources to run comparative probabilistic sensitivity analyses (PSAs) of screening strategies across three CRC models. PSAs use samples from the distributions of uncertain model parameters to propagate uncertainties to model predictions. To date, formal PSAs addressing uncertainties in all relevant parameters, that is, in the natural history and screening parameters, have not been performed for complex cancer screening models. The goal of this project is to take this a step further by using a comparative framework that includes three state-of-the-art CRC models. Funded under the National Cancer Institute’s (NCI) Cancer Intervention and Surveillance Modeling Network (CISNET) program, these CRC models were independently developed for the evaluation of interventions, with emphasis on screening, and describe CRC natural history using different underlying assumptions. These differences enable model comparisons that explore the impact of structural uncertainty, in addition to parameter uncertainty.
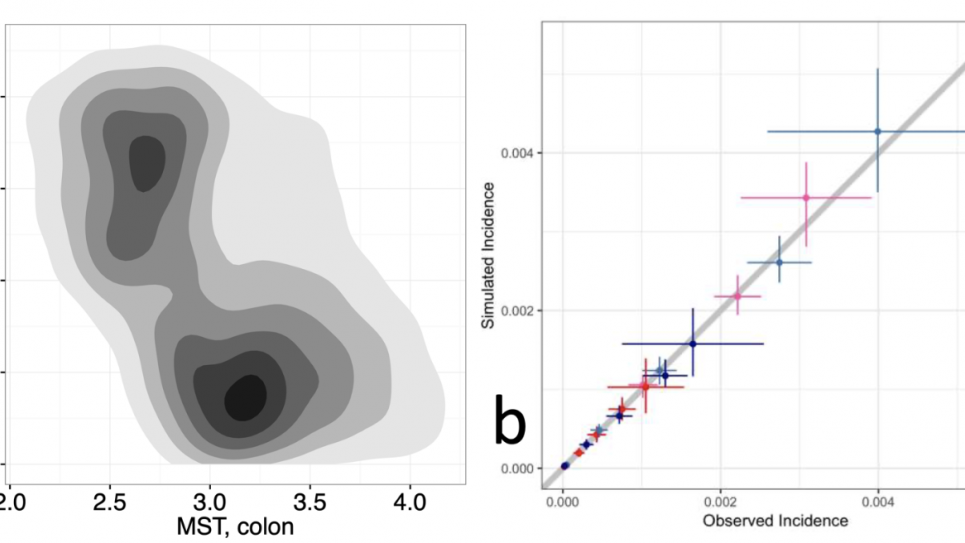
The comparative PSAs in this work will be used to generate cost-effectiveness analyses (CEAs) for complex interventions and to provide formalized assessments of both structural and parameter uncertainties across the three CRC models. For external parameters (i.e., costs, screening test characteristics and health-related quality of life), distributions will be derived from current systematic reviews. For the calibrated parameters, the joint posterior distributions for all three models will be obtained by applying sequential Bayesian calibration algorithms, using CRC registry and clinical trial data as calibration targets. The calibration algorithms will be implemented as HPC workflows using the EMEWS framework (emews.github.io) and the Swift/T workflow engine (swift-lang.org/Swift-T). EMEWS workflows have been run on leadership computing resources such as OLCF Summit, NERSC Cori and ALCF Theta. Additional EMEWS workflows will be used to sample from the joint posteriors and generate the comparative PSAs of screening strategy outcomes. This work integrates complex data, large-scale machine learning (ML) algorithms for uncertainty quantification (UQ), and simulation to advance HPC-enabled scientific discovery, supporting the DOE mission. The approaches also promote the adoption of HPC across other scientific domains as they are applicable beyond cancer or even microsimulation models to any computational modeling method (e.g., agent-based modeling) requiring heuristic/derivative free methods for UQ.