With the SARS-CoV-2 virus and associated COVID-19 disease continuing to disrupt daily life across the planet, the U.S. Department of Energy’s (DOE) Argonne National Laboratory has emerged as a powerful player in efforts to understand and combat the virus. To perform this research, various teams are leveraging Theta, a supercomputer supported by the Argonne Leadership Computing Facility (ALCF), a U.S. DOE Office of Science User Facility.
Among them is a group of researchers led by Argonne computational biologist Arvind Ramanathan, whose team aims to unravel the fundamental biological mechanisms of the virus while also identifying potential therapeutics to treat the disease. Brought together by the COVID-19 High Performance Computing Consortium, which seeks proposals from researchers around the globe, the team represents institutions from the federal government, academia, and industry.
Given the novel nature of the virus and disease, this is no straightforward task. “The virus attaches itself to the human body and it basically has a way of subverting the immune system, so we are trying to understand the underlying mechanisms which enable this subversion,” Ramanathan said.
Utilizing large-scale molecular dynamics (MD) simulations and techniques drawn from artificial intelligence (AI) and machine learning, the team has made significant progress.
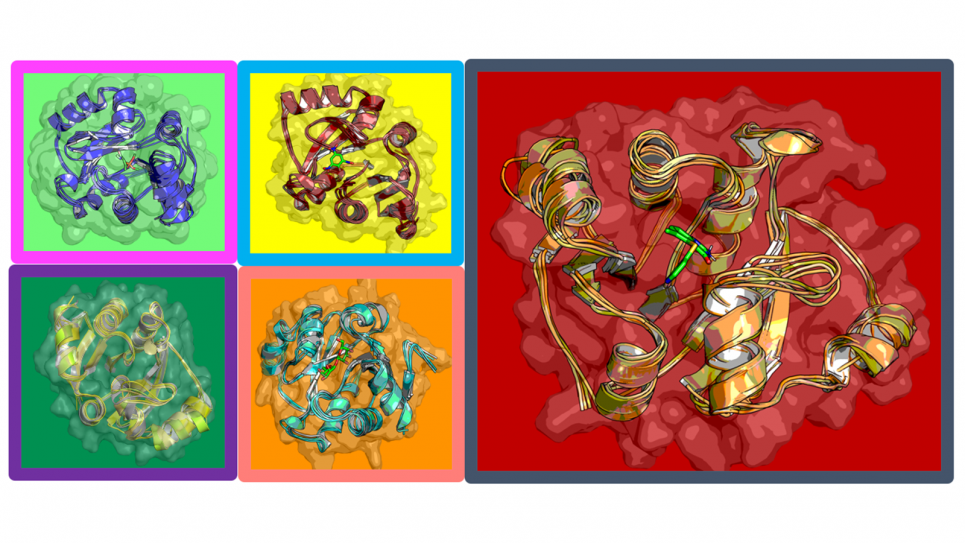
The researchers, working closely with colleagues at the Advanced Photon Source (APS), a U.S. DOE Office of Science User Facility at Argonne, use what can be described as snapshots of the virus to determine its crystal structure. From this they try and identify sites of interest—potential targets for other molecules to bind to and/or attack.
Data were taken at the Structural Biology Center (SBC) at Sector 19 of the APS, and at BioCARS at Sector 14 of the APS. The APS has dedicated more than 8,000 hours of beam time to COVID-19 research since the start of the pandemic.
“Structural biology has been a vital part of the fight against COVID-19,” said SBC Director Andrzej Joachimiak of Argonne and the University of Chicago. “Nearly 100 structures of this coronavirus’s proteins have been determined at the APS, and those structures can be used to find weaknesses in the virus’s armor.”
While the virus expresses at least 29 proteins, not all are likely targets. “Certain proteins are more important than others,” Ramanathan explained. “We focus on the ones we deem likeliest to be salient in the hope that we can identify another molecule that, upon interacting with the target protein, would prevent the virus from multiplying or otherwise behaving in ways that evoke a negative response when it affects the human body.”
Simulations are necessary for filling in details.
“In some sense, the static images we analyze are not reality,” Ramanathan cautioned. “As the snapshots fail to capture the dynamics integral to the virus, they provide an incomplete picture—a constrained view of this biomolecule, of this protein—because life is not static. And so we use simulations to understand more completely what this biomolecule, what this protein, is doing. We want to capture as much as possible.”
This includes capturing the effects of other molecules.
“When you introduce a small molecule, in some way it will modulate or change the dynamics of the protein under study, so we need to understand those modulations or changes,” which, Ramanathan noted, is like attempting to hit a moving target. The interactions that occur subsequently can help guide the researchers in predicting interesting targets on the protein.
Organically coupled with the identification of such targets in the virus is the discovery of therapeutics. Specifically, researchers in the broader group are trying to design antibodies for the virus—that is, as Ramanathan put it, “proteins that can bind to specific epitopes or regions that innately make the virus vulnerable to attack from our immune systems.”
The antibody design selection is intended to facilitate the development of a vaccine. To guide these efforts, the researchers are mining libraries of existing compounds by harnessing machine learning to rapidly search for, filter, and rank small molecules that are then virtually screened on a high-throughput basis.
To date, more than 6 million small molecules have been screened. Researchers have identified at least 20 partially active molecules with the potential to inhibit viral functions. Through experiments involving live human lung cell cultures, these candidates are being validated in Argonne laboratories for activity against the virus.
The work is computationally demanding, with the data-intensive antibody design selection process in particular being propelled by powerful supercomputing resources. The related AI-driven MD simulations are enabled not just by Theta, but also the Summit supercomputer housed at the Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science User Facility.
In addition to those offered by the ALCF and OLCF, the computing consortium behind the project leverages resources at Texas Advanced Computing Center, San Diego Supercomputing Center, and Brookhaven National Laboratory. Data from this work are publicly available.
“On top of everything, there is another side to this work: the high-performance computing angle,” Ramanathan said. “Further calamities will occur in the future, so we need to be able to study them—how can we enable our supercomputers to be ready for these types of challenges when they emerge?”
In a related effort, Ramanathan and collaborators produced a paper, "AI-Driven Multiscale Simulations Illuminate Mechanisms of SARS-CoV-2 Spike Dynamics," that has been named a finalist for the ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research. The award winner will be announced at the SC20 conference.
This research was supported by the Exascale Computing Project, a collaborative effort of the U.S. DOE Office of Science and the National Nuclear Security Administration, and the DOE’s National Virtual Biotechnology Laboratory with funding from the Coronavirus CARES Act. This work used resources, services, and support from the COVID-19 HPC Consortium. Additional ALCF computing time was provided through a Director's Discretionary allocation.
Additional funding for APS beamlines used for COVID-19 research is provided by the National Institutes of Health (NIH) and by DOE Office of Science Biological and Environmental Research. The APS operated for 10 percent more hours this year than usual to support COVID-19 research, with the additional time supported by the DOE Office of Science through the National Virtual Biotechnology Laboratory, with funding provided by the Coronavirus CARES Act.
==========
The Argonne Leadership Computing Facility provides supercomputing capabilities to the scientific and engineering community to advance fundamental discovery and understanding in a broad range of disciplines. Supported by the U.S. Department of Energy’s (DOE’s) Office of Science, Advanced Scientific Computing Research (ASCR) program, the ALCF is one of two DOE Leadership Computing Facilities in the nation dedicated to open science.
About the Advanced Photon Source
The U. S. Department of Energy Office of Science’s Advanced Photon Source (APS) at Argonne National Laboratory is one of the world’s most productive X-ray light source facilities. The APS provides high-brightness X-ray beams to a diverse community of researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. These X-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being. Each year, more than 5,000 researchers use the APS to produce over 2,000 publications detailing impactful discoveries, and solve more vital biological protein structures than users of any other X-ray light source research facility. APS scientists and engineers innovate technology that is at the heart of advancing accelerator and light-source operations. This includes the insertion devices that produce extreme-brightness X-rays prized by researchers, lenses that focus the X-rays down to a few nanometers, instrumentation that maximizes the way the X-rays interact with samples being studied, and software that gathers and manages the massive quantity of data resulting from discovery research at the APS.
This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation’s first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America’s scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy’s Office of Science.
The U.S. Department of Energy’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.